Pharmacogenetics is often defined as the study of variation in response to drugs due to hereditary factors. Variation in therapeutic efficacy or side effects experienced may be due to both pharmacokinetic and pharmacodynamic factors (1). We know most about genetic variations in drug-metabolising enzymes, particularly the cytochrome P-450 (CYP) enzymes. CYP2D6 is one of the most thoroughly studied and clinically the most important of the polymorphic CYP enzymes (2 – 4). CYP2D6 is involved in the metabolism of a large number of drugs, including many antidepressants and antipsychotics (5, 6). In consequence, pharmacogenetic testing in a clinical context is highly relevant in psychopharmacology, as shown in previous articles in this Journal (7, 8).
Null mutations in the CYP2D6 gene cause lack of enzyme activity (3, 4). Metabolism of many drugs therefore takes place more slowly in individuals who are homozygous for inactivating mutations of this kind (i.e. both CYP2D6 alleles are non-functional). These individuals are called poor metabolisers (PM). Persons with two normal copies of the CYP2D6 gene are called extensive metabolisers (homozygous EM). Enzyme activity in individuals with one inactive and one normal allele (heterozygous EM) is also usually high enough for drug metabolism to be within the range defined as normal (2). Some subjects have three or more active CYP2D6 alleles as a result of hereditary gene duplication or gene amplification, and this may cause higher enzyme activity. These individuals are called ultrarapid metabolisers (UM). There are large ethnic differences in the distribution of the various CYP2D6 genotypes (9, 10). In Northern Europe the percentage of PMs is 5 – 10 % and the percentage of UMs is 1 – 2 %. The situation in north-east Africa is the reverse: up to 30 – 40 % of the population are UMs and only 1 % are PMs. In East Asia the percentage of both PMs and UMs is low at around 1 – 2 %.
Poor metabolisers are at risk of having a higher than expected serum concentration in relation to the drug dose, and hence more side effects. Ultrarapid metabolisers, on the other hand, may have a low serum concentration in relation to the dose, and will often experience less effect from the drug (6). However, the opposite can also be seen in those cases where CYP2D6 is involved in the metabolism of an inactive «prodrug» into an active metabolite. This is true, for example, of codeine and tamoxifen (4, 11). The consequence of being a PM will then be that the drug in question will have less effect or none at all.
It is believed that the extent of unintended side effects and therapeutic failure can be reduced by adapting drug treatment to the patient’s CYP2D6 genotype, and increased use of pharmacogenetic testing in clinical practice is often recommended (2, 3, 5, 7). CYP2D6 genotyping of this kind is offered today at a number of public laboratories in Norway. Commercial operators are also very interested in seeing an increase in the amount of genotyping, and offer simultaneous testing of many genetic variants with the aid of microarray technology. However, there is still limited documentation of the cost-effectiveness of pharmacogenetic testing in a practical clinical situation (12 – 14).
Our undocumented experience gained from the routine CYP2D6 analyses indicates that they are of limited value. The purpose of this study has thus been to investigate more systematically the results of CYP2D6 genotyping in clinical practice. Theoretically, one would expect to find a considerably larger proportion of both PMs and UMs among the patients examined than in the population at large. We also wanted to explore whether the clinical indication in the requisition was consistent with and could predict the probability of finding a genotype corresponding to abnormal drug metabolism in the patients.
Materials and method
Patient material
The sample database at the Centre for Medical Genetics and Molecular Medicine at Haukeland University Hospital was used to identify retrospectively all requisitions with a query about CYP2D6 genotyping from 29 June 1998 to 28 December 2009. 328 samples were considered. 325 were included, while three were excluded because of ambiguous CYP2D6 genotyping results (Fig. 1). On the basis of an overall assessment of the information in the requisition, the clinical problem was registered as question of either poor metabolism or ultrarapid metabolism. Where the data were inadequate or inconsistent, the clinical problem was registered as «unclear». Drugs, dose and serum concentration were also recorded – in those cases where the data were available. The requisitions were then classified into three different groups on the basis of the data recorded (Table 1). The result of the genotyping was not known to the person performing the classification into groups. The classification of clinical problem and indication groups was primarily carried out by one of the authors (HHV) and the grouping was then verified by the project manager (VMS). Agreement was reached on the classification in all cases. Requisitions in Group 1 had a phenotype consistent with abnormal drug metabolism. In addition, the patients had used at least one drug where CYP2D6 is essential in the metabolism (Table 2) (6, 15, 16). Group 2 also consisted of requisitions with information about a phenotype consistent with abnormal drug metabolism, but either the drug specified is only metabolised to a limited extent or not at all by CYP2D6, or the drug was not specified. Requisitions with limited or completely contradictory information were put in Group 3.
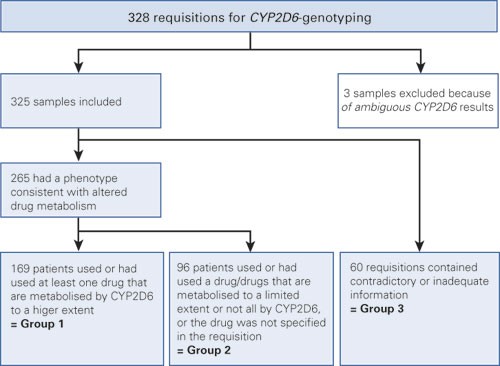
Figure 1 Schematic overview of the grouping of requisitions for CYP2D6 genotyping at the Centre for Medical Genetics and Molecular Medicine, Haukeland University Hospital, in the period 1998 – 2009.
|
Table 1 Division into indication groups, based on information on the requisition form. CYP2D6 genotyping at the Centre for Medical Genetics and Molecular Medicine, Haukeland University Hospital, in the period 1998 – 2009.
|
|
Group 1:
|
Increased side effects or lack of therapeutic effect of a CYP2D6 substrate and/or abnormal serum level
|
|
Group 2:
|
Increased side effects or lack of therapeutic effect and/or abnormal serum level. Medication not stated or not metabolised significantly by CYP2D6.
|
|
Group 3:
|
Not classifiable
|
|
Table 2 Overview of how specified drugs were placed in indication Groups 1 and 2 respectively¹
|
|
Group 1
|
Group 2
|
|
Drugs where CYP2D6 metabolism is of major importance to the clinical drug response
|
Drugs where CYP2D6 is of limited or no importance to metabolism
|
|
Amitriptyline
|
Citalopram
|
|
Aripiprazole
|
Escitalopram
|
|
Fluoxetine
|
Flupentixol
|
|
Fluvoxamine
|
Foscarnet
|
|
Haloperidol
|
Carbamazepine
|
|
Imipramine
|
Clozapine
|
|
Clomipramine
|
Quetiapine
|
|
Chlorpromazine
|
Lamotrigine
|
|
Chlorprothixene
|
Levomepromazine
|
|
Codeine²
|
Lithium
|
|
Mianserin
|
Mirtazapine
|
|
Nefazodone
|
Olanzapine
|
|
Nortriptyline
|
Sertraline
|
|
Paroxetine
|
Valproate
|
|
Perphenazine
|
Ziprasidone
|
|
Risperidone
|
|
|
Thioridazine
|
|
|
Trimipramine
|
|
|
Venlafaxine
|
|
|
Zuclopenthixol
|
|
|
[i]
|
Control group
In order to be able to estimate and make a comparison with the frequency of CYP2D6 genotypes in a Norwegian population, an anonymised control sample collected at the Centre for Medical Genetics and Molecular Medicine in 1998 was used. The control material consisted of blood samples from 100 random, healthy blood donors (50 women and 50 men) at Haukeland University Hospital.
CYP2D6 genotyping
All the samples were investigated for the PM-associated non-functional alleles CYP2D6*3 (2549delA), CYP2D6*4 (1846G>A), CYP2D6*5 (deletion of the whole CYP2D6 gene) and CYP2D6*6 (1707delT) using the methods previously described (17, 18). In a northern European population, screening for these four alleles is expected to reveal at least 95 % PMs for CYP2D6-mediated metabolism (9, 19). UM-associated duplications of the CYP2D6 gene were investigated by means of long-range PCR (20). When two non-functional alleles were detected, the genotype was denoted a PM. In persons with one non-functional allele and one normal allele, the genotype was denoted heterozygous EM. When two normal alleles were detected, the genotype was denoted homozygous EM. Persons with one non-functional allele and one duplicate allele for ultra-rapid metabolism were also classified as («homozygous») EM. One duplicate and one normal allele were assigned to the category UM.
Ethics
Regional Ethics Committee West (REK Vest) regards this project as a quality control study which it is not mandatory to submit to the committee. The project was also discussed with the Bergen Hospital Trust’s data protection officer («personvernombud»), who was of the opinion that it was not subject to reporting.
Statistics
Statistical calculations were carried out using Fisher’s exact test, and the limit value for statistical significance was set at p < 0.05.
Results
Of the 325 requisitions in the study, 169 were placed in Group 1, 96 in Group 2 and 60 in Group 3 (Fig. 1).
236 patients (72.6 %) were treated with psychopharmacological drugs. 106 (32.6 %) used or had used at least one specific antipsychotic drug, 69 (21.2 %) at least one specific antidepressant drug, 25 (7.7 %) a combination of antipsychotics and antidepressants and 36 patients (11.1 %) had not used a specified psychiatric drug. In all, 290 requisitions (90.2 %) were associated with psychiatry in that the patients were either using psychopharmacological drugs (n = 236; 72.6 %), had a psychiatric diagnosis (n = 154; 47.4 %), the test was ordered by a psychiatrist or other physician at a psychiatric institution (n = 242; 74.5 %) or a combination of these.
Allele frequencies and genotype distribution in the different groups are shown in Table 3. 4.0 % of all the patients were genotypical UMs, as compared to none in the Control Group (p = 0.045). At group level, the percentage of UMs was higher in Group 1 (5.3 %) than in the Control Group (0 %) (p = 0.029), whereas the percentages in Group 2 and Group 3 were not statistically significantly different from that of the Control Group. The proportion of PMs was 8.3 % among the patient group as a whole and 6.0 % in the Control Group (p = 0.528). Of the sub-groups, Group 1 had the highest proportion of PMs (10.7 %), but this was not statistically significantly different from the Control Group (p = 0.269). There was no statistically significant difference in the proportion of PM or UM genotypes between patients with and patients without recorded deviating serum concentration measurement (data not shown).
|
Table 3 CYP2D6 genotyping at the Centre for Medical Genetics and Molecular Medicine, Haukeland University Hospital, in the period 1998 – 2009. Distribution of patients in the various indication groups and frequency of CYP2D6 alleles and genotypes
|
|
|
Allele frequency
|
|
Genotype
|
|
Indication group
|
No. of patients
|
Non-functional allele (%)
|
Normal allele (%)
|
Duplicate allele (%)
|
|
PM
|
Heterozygous EM
|
Homozygous EM
|
UM
|
|
|
|
|
|
|
No. (%)
|
No. (%)
|
No. (%)
|
No. (%)
|
|
Group 1
|
169
|
22.8
|
74.0
|
3.3
|
|
18 (10.7)
|
39 (23.1)
|
103 (60.9)
|
9 (5.3)
|
|
Group 2
|
96
|
21.9
|
77.1
|
1.0
|
|
4 (4.2)
|
34 (35.4)
|
56 (58.3)
|
2 (2.1)
|
|
Group 3
|
60
|
25.8
|
71.7
|
2.5
|
|
5 (8.3)
|
20 (33.3)
|
33 (55.0)
|
2 (3.3)
|
|
Total
|
325
|
23.1
|
74.5
|
2.5
|
|
27 (8.3)
|
93 (28.6)
|
192 (59.1)
|
13 (4.0)
|
|
Control persons
|
100
|
28.5
|
71.0
|
0.5
|
|
6 (6.0)
|
44 (44.0)
|
50 (50.0)
|
0 (0,0)
|
Clinical indications in the requisition were a question of poor metabolism in 132 samples (40.6 %) and of ultrarapid metabolism in 136 samples (41.8 %), whereas for 57 of the samples (17.5 %) the indication for testing was unclear (Table 4a). Among the samples with a question of poor metabolism (n = 132) we found a total of 17 patients (12.9 %) with the classification genotypical PM as compared to seven patients (5.1 %) with a clinical indication of ultrarapid metabolism (p = 0.032) and 6.0 % of the control persons (p = 0.119) (Table 4a). Among samples with a question of ultrarapid metabolism (n = 136) there were seven (5.1 %) who were UMs by genotype, compared with three (2.3 %) UMs in the group of patients with assumed poor metabolism (p = 0.335) and none (0 %) of the Control Group (p = 0.022).
|
Table 4 a) Result of CYP2D6 genotyping in relation to clinical indication at the Centre for Medical Genetics and Molecular Medicine, Haukeland University Hospital, in the period 1998 – 2009.
|
|
|
Result
|
|
Group
|
Clinical indication
|
PM
|
|
Heterozygous EM
|
|
Homozygous EM
|
|
UM
|
Total
|
|
1
|
PM
|
11
|
|
|
22
|
|
|
47
|
|
|
2
|
|
82
|
|
UM
|
7
|
|
|
16
|
|
|
51
|
|
|
5
|
|
79
|
|
Unclear
|
|
|
|
1
|
|
|
5
|
|
|
2
|
|
8
|
|
2
|
PM
|
4
|
|
|
15
|
|
|
22
|
|
|
|
|
41
|
|
UM
|
|
|
|
19
|
|
|
32
|
|
|
2
|
|
53
|
|
Unclear
|
|
|
|
|
|
|
2
|
|
|
|
|
2
|
|
3
|
PM
|
2
|
|
|
2
|
|
|
4
|
|
|
1
|
|
9
|
|
UM
|
|
|
|
2
|
|
|
2
|
|
|
|
|
4
|
|
Unclear
|
3
|
|
|
16
|
|
|
27
|
|
|
1
|
|
47
|
|
Total
|
PM
|
17
|
(12.9 %)
|
|
39
|
(29.5 %)
|
|
73
|
(55.3 %)
|
|
3
|
(2.3 %)
|
132
|
|
UM
|
7
|
(5.1 %)
|
|
37
|
(27.2 %)
|
|
85
|
(62.5 %)
|
|
7
|
(5.1 %)
|
136
|
|
Unclear
|
3
|
(5.3 %)
|
|
17
|
(29.8 %)
|
|
34
|
(59.6 %)
|
|
3
|
(5.3 %)
|
57
|
|
Total
|
|
27
|
(8.3 %)
|
|
93
|
(28.6 %)
|
|
192
|
(59.1 %)
|
|
13
|
(4.0 %)
|
325
|
Using the results of the CYP2D6 genotyping as the starting point (Table 4b), it is seen that of 27 patients with the PM genotype, 17 (63.0 %) had a clinical indication of poor metabolism. This percentage was higher than in the patient group as a whole, where 132 of 325 (40.6 %) had a question of PM status (p = 0.027). Interestingly, seven (25.9 %) of the patients with the PM genotype actually had a question of ultrarapid metabolism in the requisition, and there were three patients with an unclear indication (11.1 %). Of a total of 13 patients with UM genotype, there were seven (53.8 %) with a corresponding ultrarapid metaboliser indication. This percentage was not statistically significantly higher than the percentage with ultrarapid metaboliser indication in the patient group as a whole (41.8 %). Of the remaining six patients with UM genotype, three (23.1 %) had a question of poor metaboliser status while the indication on three (23.1 %) was unclear.
|
Table 4 b) Clinical indication in relation to the different results of the CYP2D6 genotyping
|
|
Clinical indication
|
|
PM
|
|
UM
|
|
Unclear
|
|
Total
|
|
Result
|
No.
|
(%)
|
|
No.
|
(%)
|
|
No.
|
(%)
|
|
No.
|
(%)
|
|
PM
|
17
|
(63.0)
|
|
7
|
(25.9)
|
|
3
|
(11.1)
|
|
27
|
(100)
|
|
Heterozygous EM
|
39
|
(41.9)
|
|
37
|
(39.8)
|
|
17
|
(18.3)
|
|
93
|
(100)
|
|
Homozygous EM
|
73
|
(38.0)
|
|
85
|
(44.3)
|
|
34
|
(17.7)
|
|
192
|
(100)
|
|
UM
|
3
|
(23.1)
|
|
7
|
(53.8)
|
|
3
|
(23.1)
|
|
13
|
(100)
|
|
Total
|
132
|
(40.6)
|
|
136
|
(41.8)
|
|
57
|
(17.5)
|
|
325
|
(100)
|
Discussion
The distribution of CYP2D6 alleles and genotypes is as expected in a northern European population (9, 10). We find a statistically significantly higher proportion of UMs among those who had undergone CYP2D6 testing than in the Control Group. There is also a tendency for the proportion of PMs to be higher in the patient group than in the Control Group, but this difference is not statistically significant. A principle finding of our survey is thus that most patients who have undergone CYP2D6 testing as a result of clinical suspicion of altered CYP2D6 metabolism in reality had a normal CYP2D6 genotype (both heterozygous EMs and homozygous EMs). This is somewhat surprising, since the patient sample is a selected group with drug treatment problems. A possible explanation is that many factors other than CYP2D6 genotype have a strong bearing on the individual variation in drug response – this may be metabolism via other enzymes, but factors such as age, gender, body weight, liver and renal function, nutritional status, smoking and interaction with other medicines may also play a role (21). The low percentage of findings of PM and UM CYP2D6 genotypes in this patient sample from ordinary clinical practice may indicate that the use of pharmacogenetic testing is not optimal.
If we look at data from the various sub-groups, we find a non-significant tendency for the proportions of both UMs and PMs to be higher in Group 1 than Group 2, which indicates that the probability of finding abnormal CYP2D6 genotype is greatest if the patient is taking a known CYP2D6 substrate and at the same time reveals a phenotype that is consistent with deviating drug metabolism. We found no statistically significant difference in the percentage with PM and UM genotypes between patients for whom an unexpected serum concentration reading had been recorded and patients for whom it had not (data not shown).
The clinical indications for genotyping were approximately equally distributed between suspicion of poor metabolism and suspicion of ultrarapid metabolism. Taking the result of the CYP2D6 genotyping as the starting point and looking only at those who were found to have the PM genotype, there was consistency between clinical suspicion and genotyping result in 63 % of cases. In a quarter of the cases, however, there was clinical question of ultrarapid metabolism, and the finding of PM genotype was thus unexpected and apparently paradoxical. One possible explanation may be that some poor metabolisers fail to take the drug that is prescribed because they experience more side effects. The result will be a low drug serum level and therapeutic failure. Of those who were found to have an UM genotype, only a little more than half had a clear ultrarapid metaboliser indication. Thus there is relatively poor correlation between findings of abnormal CYP2D6 genotype and clinical indication.
A majority of the patients who were CYP2D6 genotyped were persons who used or had used antipsychotics and/or antidepressants, and most samples were requested by a psychiatrist or another physician at a psychiatric institution. This is consistent with international experience, where CYP2D6 testing is particularly used in connection with psychopharmacological issues (2, 3). We had also expected that CYP2D6 genotyping would be of interest in connection with codeine treatment, as CYP2D6-mediated transformation of codeine to morphine is a prerequisite for its analgesic effect. However, there were few samples with queries associated with codeine metabolism in our study, which indicates low demand or little need for testing in this group. There has also been little demand for genotyping in connection with the use of other drugs that are metabolised via CYP2D6, such as some beta blockers and antiarrhythmica (1). Nor have there been any requests in our material for CYP2D6 genotyping in connection with treatment with tamoxifen, which is normally bioactivated in vivo by CYP2D6 (4). In recent years it has been reported that breast cancer patients with lack of or reduced CYP2D6 activity may benefit less from tamoxifen treatment than individuals with normal enzyme function, but the value of CYP2D6 genotyping in this setting remains contentious (4, 22).
Non-systematic review articles have indicated that the benefit of pharmacogenetic analyses is limited to a few enzymes and drugs (21). At present, clinical genotyping of psychiatric patients is relevant for CYP2D6 and CYP2C19, and then only in cases where problems have arisen during treatment with specific drugs, such as tricyclic antidepressants and certain antipsychotics (2, 3, 21). The benefit of genotyping before the start of treatment is not documented, and it was recently questioned whether pharmacogenetics has any future at all in clinical psychiatry (23).
Nevertheless, a number of laboratories in Norway offer pharmacogenetic tests. Our data suggest that many tests are ordered without a well-founded indication for the test in question. For CYP2D6 genotyping the requisition should at least provide specific information about the drug treatment in question and the patient should also use a drug where the CYP2D6 enzyme plays a central role in the metabolism. If the knowledge emerging from pharmacogenetic research is to be of value, genotyping must be geared to a greater extent to the everyday clinical situation. We believe that the laboratories have a responsibility to pave the way for the physicians, for example by providing courses and internet-based information, so that genotyping is used in cases where it can be expected to be useful and avoided in cases where the value can be assumed a priori to be low. In a previous article in this Journal it was proposed establishing a professional body for laboratory medicine in the specialist health service (24). It is conceivable that such a body could also contribute to ensuring more correct use of pharmacogenetic analyses.
Weaknesses
One weakness of our study is that the number of patients is rather limited, particularly with respect to sub-grouping of data, which results in a risk of a type 2 errors (false negative results). The division into groups was based on existing information in the requisition and in many cases this was very limited. We were also often dependent on the physician’s assessment of «abnormal» serum level, but without access to the underlying clinical data. It should be noted that assessments of this kind must take account of both the absolute serum drug concentration compared with the recommended therapeutic window and the ratio between drug dose and serum level. At Haukeland University Hospital there is no established practice for measuring the serum concentration of a drug prior to CYP2D6 genotyping. However, we have received samples from all over Norway, and the significance of prior therapeutic drug monitoring was not apparent in our material. It is possible, nevertheless, that a higher proportion of PMs and UMs would be found if genotyping took place only in the light of abnormal findings in connection with drug serum concentration measurements. Finally, it is a weakness that we do not know whether the result of CYP2D6 genotyping influenced future treatment of the patient. We have not obtained permission to contact the physicians subsequently, so we do not know the real clinical value of the genotyping.
Conclusion
Our survey shows that CYP2D6 genotyping in ordinary clinical practice results in a relatively low percentage of PM and UM findings, even though the result of the genetic testing has clearly yielded some useful answers for individual patients. The main problems appear to be an inadequately documented indication, with requests for genotyping even when the drug is only metabolised to a limited extent by CYP2D6, and that a number of different non-genetic factors may influence the drug response.